
The confluence of digital transformation in biomedicine and the current generative AI revolution creates an unprecedented opportunity for drastically accelerating progress in precision health. Digital pathology is emblematic of this exciting frontier. In cancer care, whole-slide imaging has become routinely available, which transforms a microscopy slide of tumor tissue into a high-resolution digital image. Such whole-slide images contain key information for deciphering the tumor microenvironment, which is critical for precision immunotherapy (for example differentiating hot versus cold tumors based on lymphocyte infiltration). Digital pathology can also be combined with other multimodal, longitudinal patient information in multimodal generative AI for scaling population-level, real-world evidence generation.
This is an exciting time, tempered by the reality that digital pathology poses unique computational challenges, as a standard gigapixel slide may be thousands of times larger than typical natural images in both width and length. Conventional vision transformers struggle with such an enormous size as computation for self-attention grows dramatically with the input length. Consequently, prior work in digital pathology often ignores the intricate interdependencies across image tiles in each slide, thus missing important slide-level context for key applications such as modeling the tumor microenvironment.
In this blog post, we introduce GigaPath (opens in new tab), a novel vision transformer that attains whole-slide modeling by leveraging dilated self-attention to keep computation tractable. In joint work with Providence Health System and the University of Washington, we have developed Prov-GigaPath (opens in new tab), an open-access whole-slide pathology foundation model pretrained on more than one billion 256 X 256 pathology images tiles in more than 170,000 whole slides from real-world data at Providence. All computation was conducted within Providence’s private tenant, approved by Providence Institutional Review Board (IRB).
To our knowledge, this is the first whole-slide foundation model for digital pathology with large-scale pretraining on real-world data. Prov-GigaPath attains state-of-the-art performance on standard cancer classification and pathomics tasks, as well as vision-language tasks. This demonstrates the importance of whole-slide modeling on large-scale real-world data and opens new possibilities to advance patient care and accelerate clinical discovery.
Spotlight: Blog post
Eureka: Evaluating and understanding progress in AI
How can we rigorously evaluate and understand state-of-the-art progress in AI? Eureka is an open-source framework for standardizing evaluations of large foundation models, beyond single-score reporting and rankings. Learn more about the extended findings.
Adapting dilated attention and LongNet to digital pathology

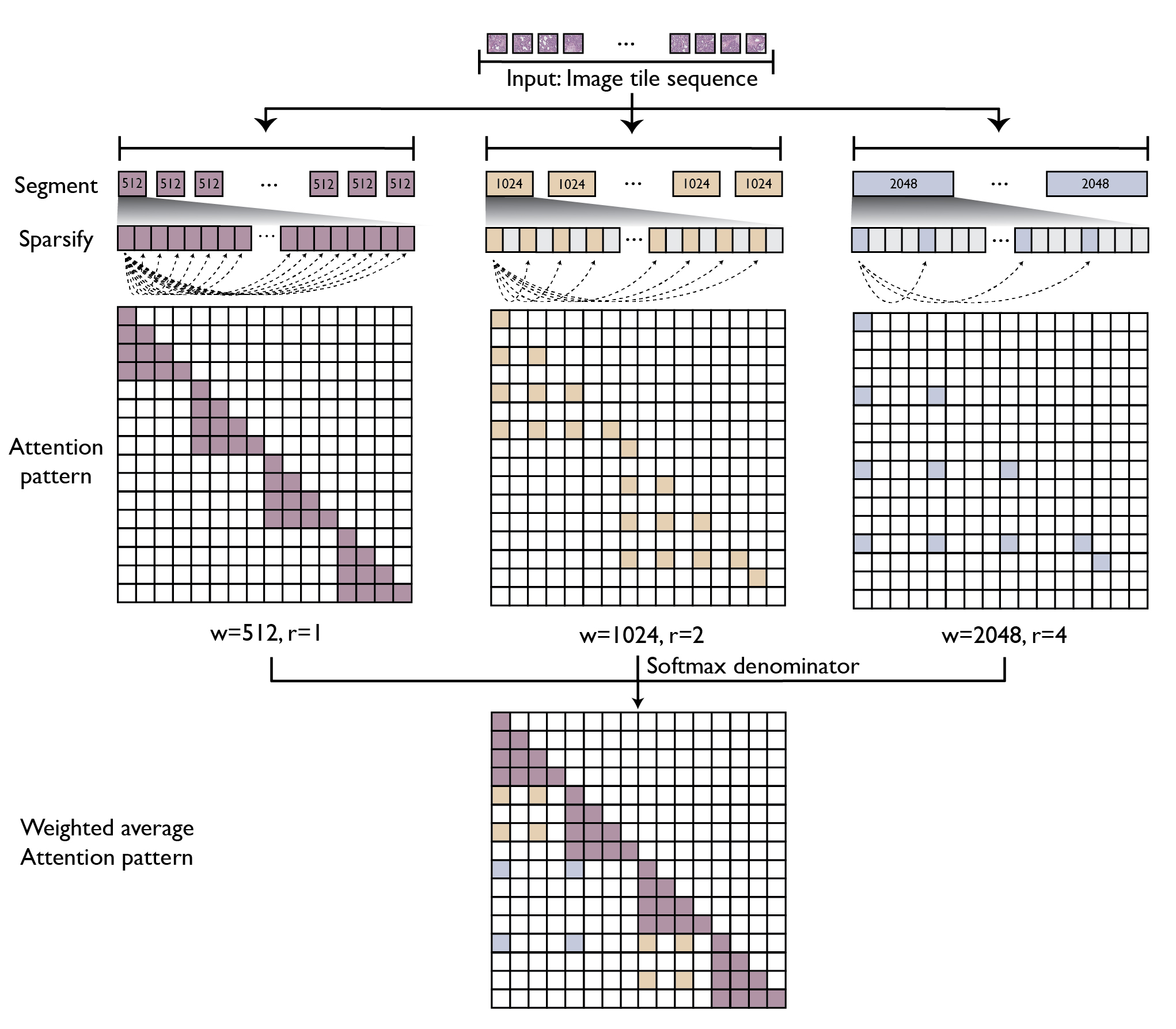
GigaPath adopts two-stage curriculum learning comprising tile-level pretraining using DINOv2 and slide-level pretraining using masked autoencoder with LongNet (see Figure 1). DINOv2 is a standard self-supervision method that combines contrastive loss and masked reconstruction loss in training teacher and student vision transformers. However, due to the computational challenge for self-attention, its application is limited to small images such as 256 × 256 tiles. For slide-level modeling, we adapt dilated attention from LongNet to digital pathology (see Figure 2). To handle the long sequence of image tiles for a whole slide, we introduce a series of increasing sizes for subdividing the tile sequence into segments of the given size. For larger segments, we introduce sparse attention with sparsity proportional to segment length, thus canceling out the quadratic growth. The largest segment would cover the entire slide, though with sparsely subsampled self-attention. This enables us to capture long-range dependencies in a systematic way while maintaining tractability in computation (linear in context length).
GigaPath on cancer classification and pathomics tasks
We construct a digital pathology benchmark comprising nine cancer subtyping tasks and 17 pathomics tasks, using both Providence and TCGA data. With large-scale pretraining and whole-slide modeling, Prov-GigaPath attains state-of-the-art performance on 25 out of 26 tasks, with significant improvement over the second-best model on 18 tasks.

On cancer subtyping, the goal is to classify fine-grained subtypes based on the pathology slide. For example, for ovarian cancer, the model needs to differentiate among six subtypes: Clear Cell Ovarian Cancer, Endometrioid Ovarian Cancer, High-Grade Serous Ovarian Cancer, Low-Grade Serous Ovarian Cancer, Mucinous Ovarian Cancer, and Ovarian Carcinosarcoma. Prov-GigaPath attained state-of-the-art performance in all nine tasks, with significant improvement over the second best in six out of nine tasks (see Figure 3). For six cancer types (breast, kidney, liver, brain, ovarian, central nervous system), Prov-GigaPath attains an AUROC of 90% or higher. This bodes well for downstream applications in precision health such as cancer diagnostics and prognostics.
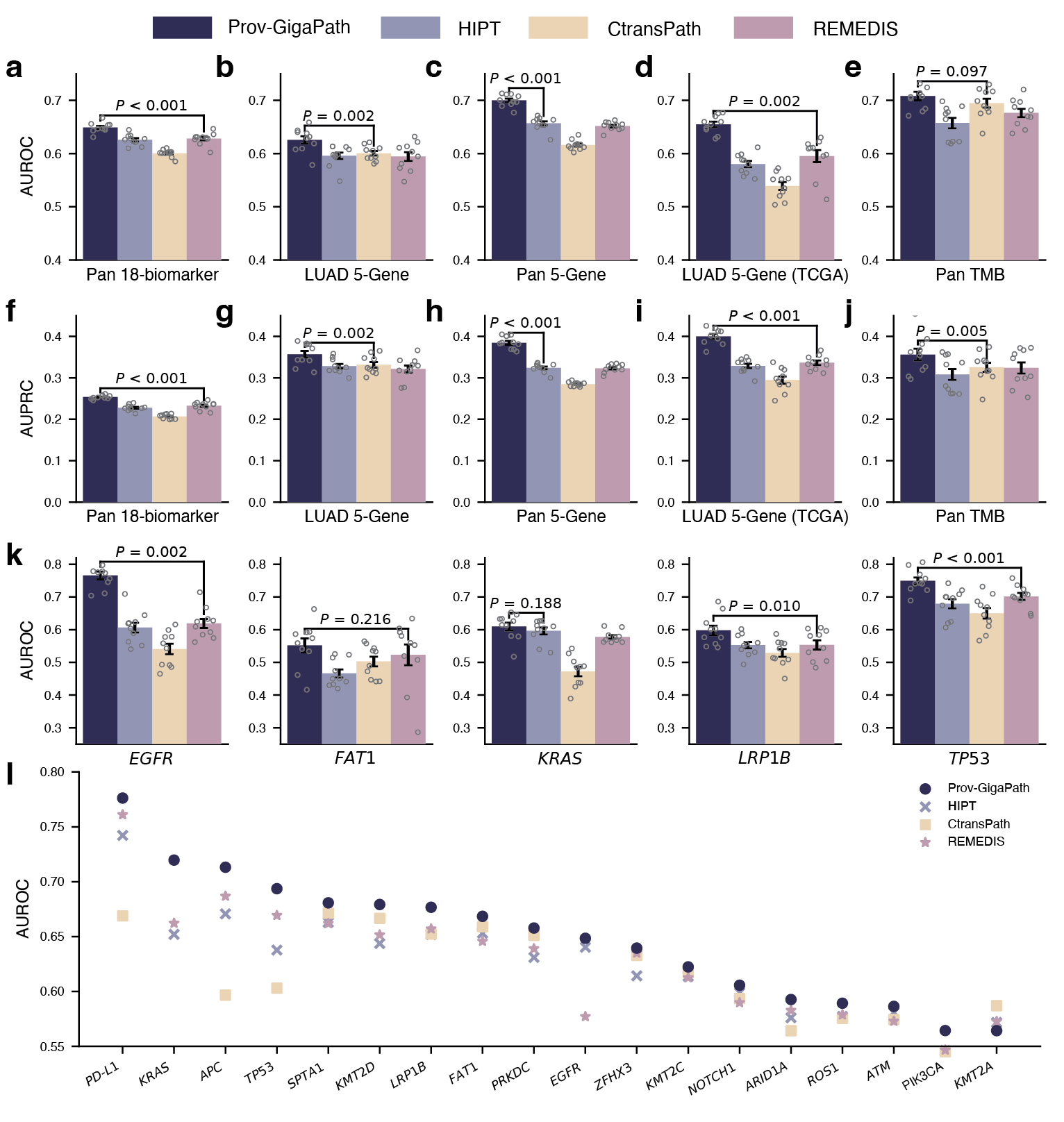
On pathomics tasks, the goal is to classify whether the tumor exhibits specific clinically relevant genetic mutations based on the slide image alone. This may uncover meaningful connections between tissue morphology and genetic pathways that are too subtle to be picked up by human observation. Aside from a few well-known pairs of specific cancer type and gene mutations, it is unclear how much signal there exists from the slide alone. Moreover, in some experiments, we consider the pan-cancer scenario, where we are trying to identify universal signals for a gene mutation across all cancer types and very diverse tumor morphologies. In such challenging scenarios, Prov-GigaPath once again attained state-of-the-art performance in 17 out of 18 tasks, significantly outperforming the second best in 12 out of 18 tasks (see Figure 4). For example, in the pan-cancer 5-gene analysis, Prov-GigaPath outperformed the best competing methods by 6.5% in AUROC and 18.7% in AUPRC. We also conducted head-to-head comparison on TCGA data to assess the generalizability of Prov-GigaPath and found that Prov-GigaPath similarly outperformed all competing methods there. This is all the more remarkable given that the competing methods were all pretrained on TCGA. That Prov-Gigapath can extract genetically linked pan-cancer and subtype-specific morphological features at the whole-slide level highlights the biological relevance of the underlying learned embeddings, and opens the door to using real-world data for future research directions around the complex biology of the tumor microenvironment.
GigaPath on vision-language tasks
![Figure 5: Comparison on vision-language tasks. a, Flow chart showing the fine-tuning of Prov-GigaPath using pathology reports. Real-world pathology reports are processed using GPT-3.5 from OpenAI to remove information irrelevant to cancer diagnosis. We performed the CLIP-based contrastive learning to align Prov-GigaPath and PubMedBERT. b, The fine-tuned Prov[1]GigaPath can then be used to perform zero-shot cancer subtyping and mutation prediction. The input of Prov-GigaPath is a sequence of tiles segmented from a WSI, and the inputs of the text encoder PubMedBERT are manually designed prompts representing cancer types and mutations. Based on the output of Prov-GigaPath and PubMedBERT, we can calculate the probability of the input WSI being classified into specific cancer subtypes and mutations. c, Bar plots comparing zero-shot subtyping performance on NSCLC and COADREAD in terms of BACC, precision and f 1. d, Bar plots comparing the performance on mutation prediction using the fine-tuned model for six genes. c,d, Data are mean ± s.e.m. across n = 50 experiments. The listed P value indicates the significance for Prov-GigaPath outperforming the best comparison approach, with one-sided Wilcoxon test. e, Scatter plots comparing the performance between Prov-GigaPath and MI-Zero in terms of BACC on zero-shot cancer subtyping. Each dot indicates one trial with a particular set of text query formulations.](https://www.microsoft.com/en-us/research/uploads/prod/2024/05/Figure-5-color-adjust.jpg)
We further demonstrate the potential of GigaPath on vision-language tasks by incorporating the pathology reports. Prior work on pathology vision-language pretraining tends to focus on small images at the tile level. We instead explore slide-level vision-language pretraining. By continuing pretraining on slide-report pairs, we can leverage the report semantics to align the pathology slide representation, which can be used for downstream prediction tasks without supervised fine-tuning (e.g., zero-shot subtyping). Specifically, we use Prov-GigaPath as the whole-slide image encoder and PubMedBERT as the text encoder, and conduct contrastive learning using the slide-report pairs. This is considerably more challenging than traditional vision-language pretraining, as we do not have fine-grained alignment information between individual image tiles and text snippets. Prov-GigaPath substantially outperforms three state-of-the-art pathology vision-language models in standard vision-language tasks, such as zero-shot cancer subtyping and gene mutation prediction, demonstrating the potential for Prov-GigaPath in whole-slide vision-language modeling (see Figure 5).
GigaPath is a promising step toward multimodal generative AI for precision health
We have conducted thorough ablation studies to establish the best practices in whole-slide pretraining and vision-language modeling. We also observed early indications of the scaling law in digital pathology, where larger-scale pretraining generally improved downstream performance, although our experiments were still limited due to computational constraints.
Going forward, there are many opportunities for progress. Prov-GigaPath attained state-of-the-art performance compared to prior best models, but there is still significant growth space in many downstream tasks. While we have conducted initial exploration on pathology vision-language pretraining, there is still a long way to go to pursue the potential of a multimodal conversational assistant, specifically by incorporating advanced multimodal frameworks such as LLaVA-Med (opens in new tab). Most importantly, we have yet to explore the impact of GigaPath and whole-slide pretraining in many key precision health tasks such as modeling tumor microenvironment and predicting treatment response.
GigaPath is joint work with Providence Health System and the University of Washington’s Paul G. Allen School of Computer Science & Engineering, and brings collaboration from multiple teams within Microsoft*. It reflects Microsoft’s larger commitment on advancing multimodal generative AI for precision health, with exciting progress in other digital pathology research collaborations such as Cyted (opens in new tab), Volastra (opens in new tab), and Paige (opens in new tab) as well as other technical advances such as BiomedCLIP (opens in new tab), LLaVA-Rad (opens in new tab), BiomedJourney (opens in new tab), BiomedParse (opens in new tab), MAIRA (opens in new tab), Rad-DINO (opens in new tab), Virchow (opens in new tab).
(Acknowledgment footnote) *: Within Microsoft, it is a wonderful collaboration among Health Futures, MSRA, MSR Deep Learning, and Nuance.
Paper co-authors: Hanwen Xu, Naoto Usuyama, Jaspreet Bagga, Sheng Zhang, Rajesh Rao, Tristan Naumann, Cliff Wong, Zelalem Gero, Javier Gonz ́alez, Yu Gu, Yanbo Xu, Mu Wei, Wenhui Wang, Shuming Ma, Furu Wei, Jianwei Yang, Chunyuan Li, Jianfeng Gao, Jaylen Rosemon, Tucker Bower, Soohee Lee, Roshanthi Weerasinghe, Bill J. Wright, Ari Robicsek, Brian Piening, Carlo Bifulco, Sheng Wang, Hoifung Poon.





