Widespread Impact of HLA Restriction on Immune Control and Escape Pathways in HIV-1
- Jonathan M. Carlson ,
- Jennifer Listgarten ,
- Nico Pfeifer ,
- Vincent Tan ,
- Carl Kadie ,
- Bruce D. Walker ,
- Thumbi Ndung'u ,
- Roger Shapiro ,
- John Frater ,
- Zabrina L. Brumme ,
- Philip J. R. Goulder ,
- David Heckerman
Journal of Virology | , Vol 86(9): pp. 5230-5243
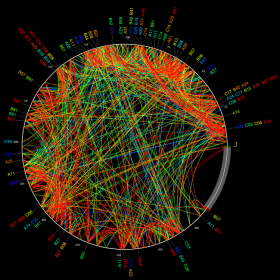
Widespread impact of differential escape
 This paper (opens in new tab) marks the development and introduction of our phylogenetically corrected logistic regression algorithm. This allows us to do all the standard logistic regression analyses–test for differential effects or measure effect size–as logistic regression, but do it while correcting for phylogenetic structure. You can use the tool yourself here (opens in new tab), though we have to limit to single analyses. If you’d like an executable version of the code, email me. We’re working on a better, scalable solution, so stay tuned.
This paper (opens in new tab) marks the development and introduction of our phylogenetically corrected logistic regression algorithm. This allows us to do all the standard logistic regression analyses–test for differential effects or measure effect size–as logistic regression, but do it while correcting for phylogenetic structure. You can use the tool yourself here (opens in new tab), though we have to limit to single analyses. If you’d like an executable version of the code, email me. We’re working on a better, scalable solution, so stay tuned.
 We used this approach to look at an interesting phenomenon: although we like to group HLA alleles by the their tendency to bind similar epitopes, we find that, in vivo, the escapes that evolution selects for differ by HLA. For example, when B*57:03 and B*57:02 (two very similar HLA alleles) present the same epitope, the observed escape mutations are usually different. Very surprising indeed, as it forces us to think more carefully about how (and if) we group alleles, as well as what the role of differential escape is. This work was in collaboration with Philip Goulder, John Frater, Roger Shapiro and Thumbi Ndung’u and thier labs.
We used this approach to look at an interesting phenomenon: although we like to group HLA alleles by the their tendency to bind similar epitopes, we find that, in vivo, the escapes that evolution selects for differ by HLA. For example, when B*57:03 and B*57:02 (two very similar HLA alleles) present the same epitope, the observed escape mutations are usually different. Very surprising indeed, as it forces us to think more carefully about how (and if) we group alleles, as well as what the role of differential escape is. This work was in collaboration with Philip Goulder, John Frater, Roger Shapiro and Thumbi Ndung’u and thier labs.
Abstract
The promiscuous presentation of epitopes by similar HLA class I alleles holds promise for a universal T-cell based HIV-1 vaccine. However, in some instances CTL restricted by HLA alleles with similar or identical binding motifs are known to target epitopes at different frequencies, with different functional avidities and with different apparent clinical outcomes. Such differences may be illuminated by the association of similar HLA alleles with distinctive escape pathways. Using a novel computational method featuring phylogenetically-corrected odds ratios, we systematically analyzed differential patterns of immune escape across all optimally defined epitopes in Gag, Pol and Nef in 2,126 HIV-1 clade C infected adults. Overall, we identified 301 polymorphisms in 90 epitopes associated with HLA alleles belonging to shared supertypes. We detected differential escape in 37 of 38 epitopes restricted by more than one allele, which included 278 instances of differential escape at the polymorphism level. The majority (66-97%) of these resulted from the selection of unique HLA-specific polymorphisms rather than differential epitope targeting rates, as confirmed by IFN-γ Elispot data. Discordant associations between HLA alleles and viral load were frequently observed between allele pairs that selected for differential escape. Furthermore, the total number of associated polymorphisms strongly correlated with average viral load. These studies confirm that differential escape is a widespread phenomenon and may be the norm when two alleles present the same epitope. Given the clinical correlates of immune escape, such heterogeneity suggests that certain epitopes will lead to discordant outcomes if applied universally in a vaccine.
Publication Downloads
PhyloD
March 25, 2016
Machine learning tools for modeling viral adaptation to host immune responses.
